BAFinder: A software for unknown bile acid identification using accurate mass LC-MS/MS in positive and negative modes
Yan Ma,* Yang Cao, Xiaocui Song, Yuanying Zhang, Jing Li, Yankai Wang, Xiaoqing Wu, and Xiangbing Qi
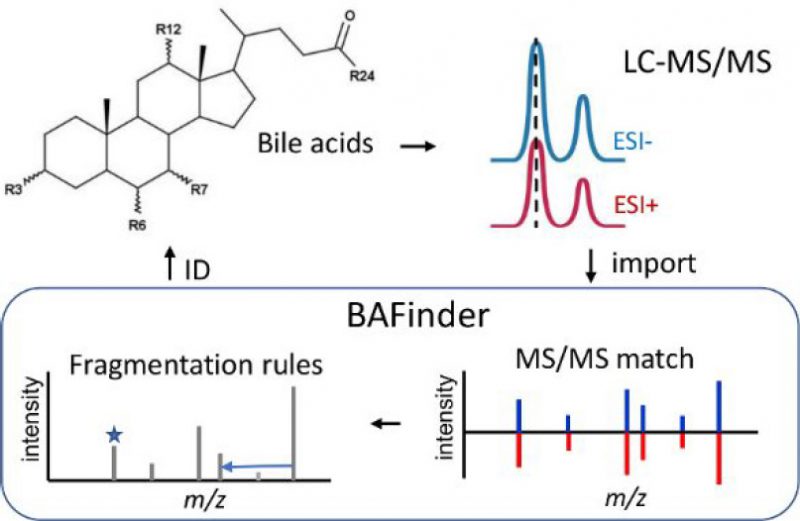
Most LC-MS based bile acid analyses target common bile acids. The identification of unknown bile acids remains challenging in untargeted experiments. Here, a software named BAFinder was developed to improve the identification of unknown bile acids from accurate mass LC-MS/MS data in both the positive and negative ESI modes. A wide variety of bile acid structures were covered in BAFinder, including oxidized bile acids and sugar conjugates that were often ignored. The annotation of unknown bile acids was based on a thorough investigation of MS/MS fragmentation patterns of 84 bile acid reference standards in both modes. Specifically, BAFinder took the peak alignment result and MS/MS spectra, grouped candidate features in positive and negative modes, searched their representative MS/MS spectra against a MS/MS library, and used characteristic product ions and neutral losses to annotate bile acids not covered in the library. Finally, the number of hydroxyl groups and double bonds, conjugation, and isomer information of bile acids were reported with four different levels of annotation confidence. The use of BAFinder was demonstrated through successful application to the analysis of human plasma and urine samples, in which a total of 112 and 244 bile acids were annotated and 75 and 111 of them were confirmed with standards or synthesized compounds, respectively. The software is freely available at https://bafinder.github.io/.